EGA Schemas
In this page, you will find information on how the European Genome-phenome Archive (EGA) manages its metadata standards using both XML Schema Definition (XSD) and JavaScript Object Notation (JSON) formats. If you are not sure what this means, you may want to explore our brief metadata introduction.
This information may be of your interest if you are planning to learn more about how the EGA is built and how to wrap around it for other processes. Nevertheless, if you are a common user (e.g. submitter or requester), you would not have to worry about these schemas nor their format, since they are implemented in user-friendly ways for you.
Metadata standards are rules that define how to format and structure data of metadata objects (i.e. entities), like EGA's samples or experiments in a consistent manner. These objects are the nodes of the metadata model of the EGA (Figure 1).
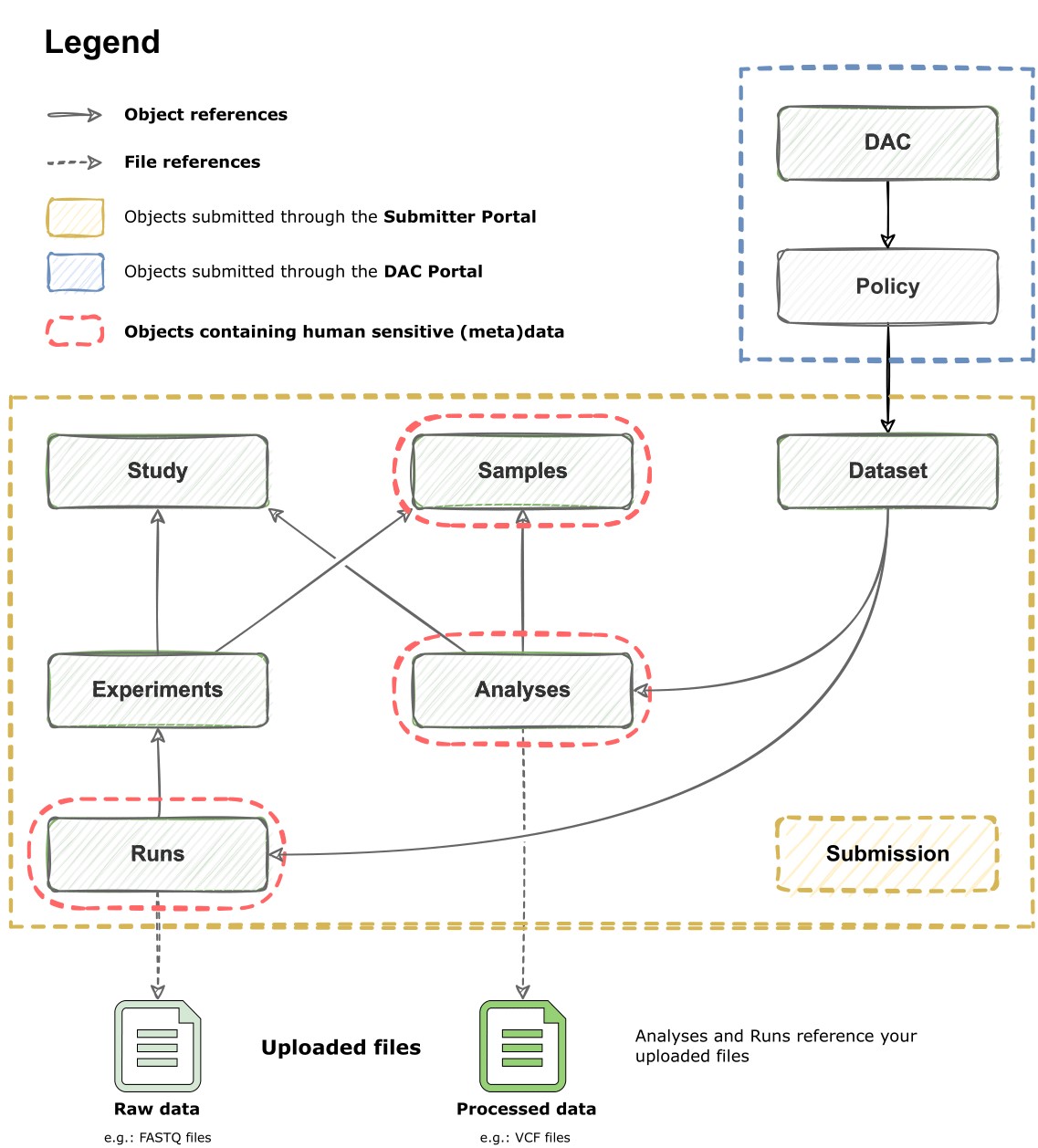
Figure 1. Diagram of EGA's metadata model. The model's building blocks are objects (e.g. sample), which can reference each other (e.g. an experiment referencing the used samples). Once your files are uploaded, they can also be referenced by Runs and Analyses. The submission object is an object itself that compiles many others.
At EGA, we inherit our metadata schemas from the European Nucleotide Archive (ENA), and we have expanded them to include bespoke objects such as "Policy", "Dataset", and "DAC" (Data Access Committee) for our specific use-case: handling sensitive human data. See below a list of all our metadata objects and some context for each.
| Metadata Object | EGA accession | Description | Examples of metadata fields |
| Study | EGAS… | Information about the study | Study type, study title, study abstract… |
| Sample | EGAN… | Information about the used samples in the experiment or analysis | Taxon ID, scientific name, biological sex, phenotype… |
| Experiment | EGAX… | Information about the performed experiment | Used libraries, sequencing platform, reference to the used samples… |
| Analysis | EGAZ… | Contains information about the analysis | Type of analysis, used assembly, reference sequence… |
| Run | EGAR… | The run holds information about the files containing the raw reads generated in a run of sequencing | Platform, spot descriptor, raw file references… |
| DAC | EGAC… | Contains information about the Data Access Committee (DAC) | DAC contacts, contact emails… |
| Policy | EGAP… | Contains the Data Access Agreement (DAA) and policy which its usage complies with | Policy text, data use ontologies (DUO) codes… |
| Dataset | EGAD… | Contains the collection of runs/analysis to be subject to controlled access | Dataset type, compilation of Run's and Analysis' IDs |
| Submission | EGAB… | Contains the collection of metadata objects that conform your whole set, as well as information about what to do with them (e.g. validate, submit, modify…) | Submission actions, metadata XML files… |
There are two different sets of schemas, based on their formats, in which the EGA accepts metadata: XSDs (for XML files) and JSON Schemas (for JSON files).
- EGA's XML Schema Definition . When programmatic submissions are pushed through the European Bioinformatics Institute (EBI) system, XML format is used. The schemas that are applied for this format are defined in XML Schema Definition (XSD) files, which can be found at ENA's GitHub repository. You can find more information on how to validate and submit your data programmatically in our programmatic submission documentation. Furthermore, see at our GitHub repository some XML examples with either made-up values (similar to what you would submit) or descriptive values for each field (just for documentation).
- EGA's JSON Metadata Schemas . When programmatic submissions are pushed through the Centre for Genomic Regulation (CRG) system, JSON format specifications are used instead. See the full JSON specifications for further details.
In conclusion, the EGA metadata schemas are crucial for maintaining the quality and consistency of submitted data. By understanding and following the rules outlined in these schemas, you can ensure that your submissions comply with the EGA's standards and contribute to a valuable and accessible genomic resource.
Sample checklists
Besides the standards in our schemas, we have another layer called Sample checklists. These are another system that the EGA inherits from ENA, and specifies what attributes are required or allowed for a sample object.
The EGA uses these checklists to enforce that, for example, a sample object has the three mandatory attributes: subject ID, sex and phenotype of the individual the sample was taken from. When you submit to EGA, our checklist is automatically selected by default.
