Submitter Portal API
The metadata submission process can be difficult and time-consuming. For this reason, the EGA has developed the Submitter Portal, a tool that was created to offer a simplified and user-friendly method of registering metadata.
Our portal provides features that are intended to make the input of metadata easier, ensuring that your data is registered correctly and effectively. Our page is divided into logical sections and includes a helpful video instruction to further assist you as you complete the submission process. With the Submitter Portal, you can rest assured that the submission of your data is in good hands.
For those that want a more flexible and automated approach, we also provide a programmatic approach using the Submitter Portal AP in addition to the user interface of the Submitter Portal. With the help of our API, you can quickly include the submission of metadata into your own workflow for a more effective and individualised experience.
Previous steps
Create your EGA account
To submit data you first need to create your EGA user. Then, once your account has been verified, you will have to request a submitter role and sign the EGA Data Processing Agreement (DPA). When the DPA is signed, you must send it to EGA Helpdesk for further validation.
Please, note that if you already have an EGA account, or you have an ega-box (submission account), you can skip this step.
Upload your files
Please note that all your files must be encrypted using the Crypt4GH tool before upload them.
As soon as you are assigned with a submitter role, you will be able to connect to the EGA inbox and upload your files.
Understand the EGA metadata schema
It's crucial to comprehend the EGA metadata schema, a set of rules that specify how data is organised, described, and shared inside the EGA, in order to get the most out of this resource. Learn all about EGA metadata schema!
Register your DAC and policy
There are two objects in the EGA metadata schema that are registered in a separate portal. All DACs and policy objects are registered using the DAC Portal, a tool developed by EGA to help data controllers manage their data stored at the EGA. You can find the relevant information in the DAC Portal Guide.
Programmatic submission
All the calls to the API need to be Authenticated. We use the OpenID Connect protocol.
API Usage Flow:
Click here to check the API usage flow schema
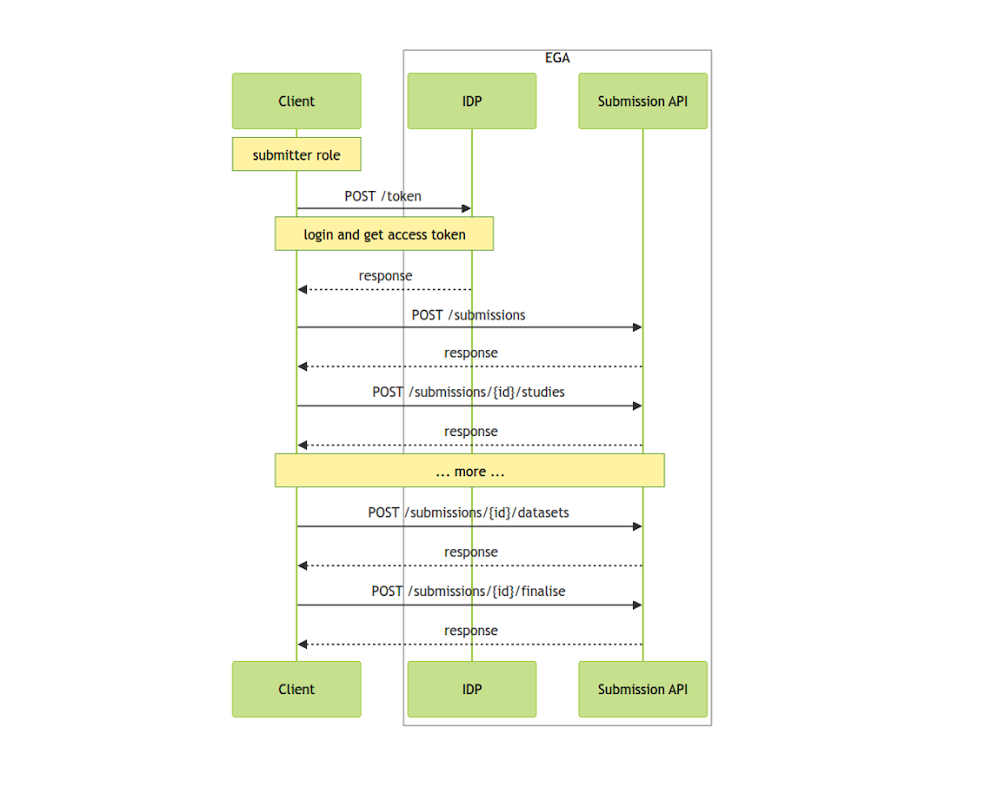
- Obtain Access and Refresh Token:
- Use Access Token in API Calls:
- Create Submission Object:
- Create EGA Metadata Objects:
- Enums
- Obtain IDs of files
- Finalise Submission:
The first step to using the API is to obtain an access and refresh token. These tokens are required for authentication and authorisation of API requests. To obtain these tokens, you need to log in using your EGA credentials.
Example:curl "https://idp.ega-archive.org/realms/EGA/protocol/openid-connect/token" \ -d "grant_type=password" \ -d "client_id=sp-api" \ -d "username=your-username" \ -d "password=your-password"
Once you have obtained the access token, you must use it in all the calls to the API. The access token is valid for a limited time period. When it expires, you will need to use the refresh token to obtain a new access token.
After authentication, you can start creating a submission object. This object will be used to store all the metadata objects and files related to the submission.
Example:curl "https://submission.ega-archive.org/api/submissions" \
-H "Authorization: Bearer your-access-token" \
-H "Content-Type: application/json" \
-d "{ \"title\":\"My submission title\",
\"description\":\"My submission description\" }"
Within the submission object, you can create other metadata objects required for the submission. These objects may include information about the submitter, study, sample, experiment, and run.
Example of study creation:curl "https://submission.ega-archive.org/api/submissions/{submisison-id}/studies" \
-H "Authorization: Bearer your-access-token" \
-H "Content-Type: application/json" \
-d "{ \"title\":\"My study title\",
\"description\":\"My study description\",
\"study_type\":\"Metagenomics\" }"
If you are reusing submitted objects, and they already have an accession, you must always use their accession. For example, when creating a new run which is reusing a previously submitted experiment, instead of using experiment_provisional_id, you must use experiment_accession_id.
There are some fields that only accept specific values, i.e. study_type. The API provides several enumerations to list these values. You can list all the enumerations, or a specific one, for example, study_types.
You can list the files available in your inbox and filter by a prefix by adding the prefix param, i.e. &prefix=/my_folder/abc
Example:curl "https://submission.ega-archive.org/api/files?status=inbox&prefix=/my_folder/abc" \ -H "Authorization: Bearer your-access-token" \ -H "Content-Type: application/json"
Once all the metadata objects are created and linked to the submission, you can finalise the submission. Finalising the submission sends it to the Helpdesk team for review.
Please, note that all files linked to the submission must be ingested, and all objects created in a submission must be linked before you can finalise the submission.
API Reference:
For a full API reference, please check our specification documentation.
