Submitting array based metadata
For further information please check our Submission FAQs, submission quickguide as well as submission terms!
The submission metadata required for Array-based submission must be submitted
using
EGA programmatic submission
and by completing the Array-based format (AF) template. The guidelines for
this workflow are described on this page.
Please notice that all files should be encrypted and uploaded prior to the
processing of your EGA Array-based-Format (AF) template.
Please notice that all files should be encrypted and uploaded prior to the processing of your EGA-Array-based-Format (AF) template.
Metadata Model
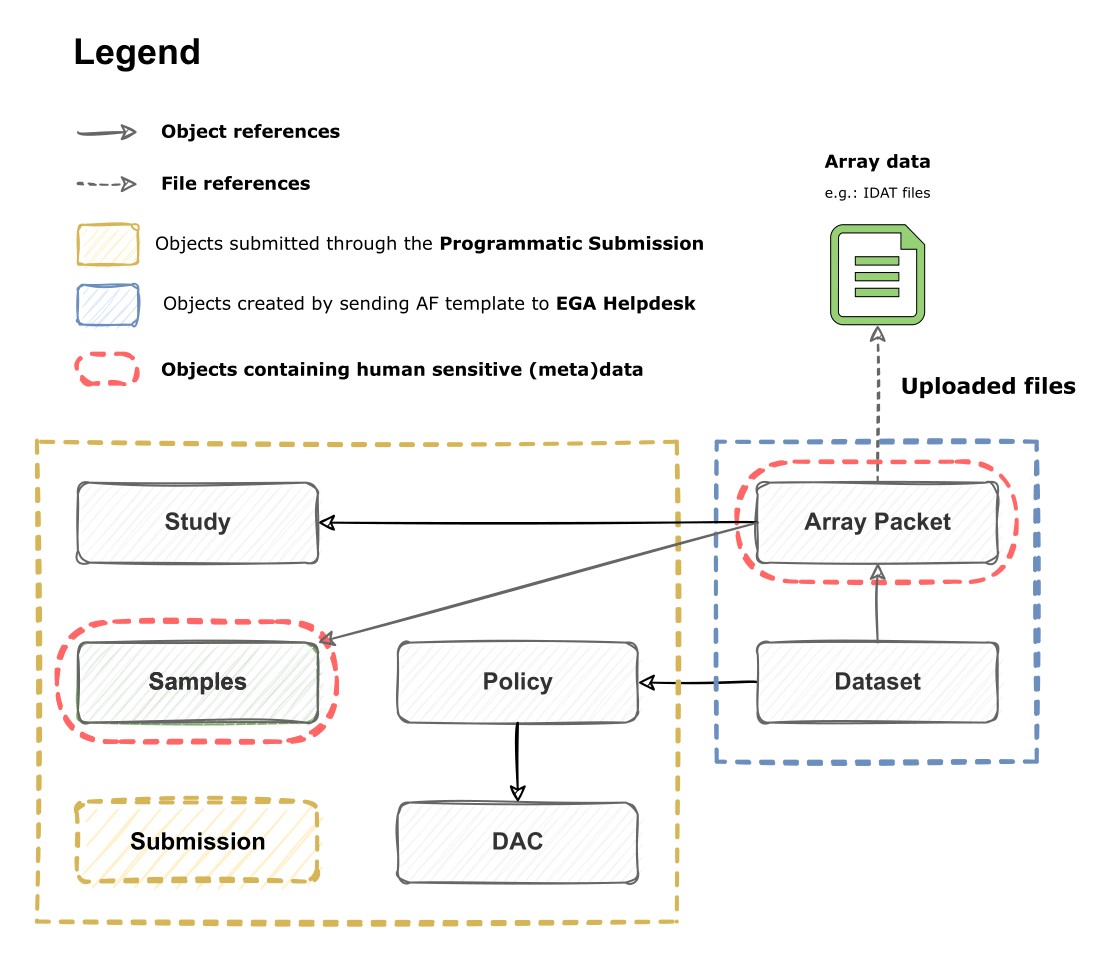
Registering Metadata
Use the EGA programmatic submission to register your Study, Samples, Data Access Committee (DAC) and Policy. This online interface enables you to create new and edit existing submissions.
Do not use the Submitter Portal to register the metadata objects for array submissions. Please, go to the EGA programmatic submission.
Registering Study
- Go to the EGA programmatic submission
- Submit an study XML
- Take a note of your study accession number (EGASXXXXXXXXXXX)
To use the study accession number in a publication, we suggest the following format:
"Sequence data has been deposited at the European Genome-phenome Archive (EGA), which is hosted by the EBI and the CRG, under accession number EGASXXXXXXXXXXX. Further information about EGA can be found on https://ega-archive.org "The European Genome-phenome Archive of human data consented for biomedical research"(https://doi.org/10.1093/nar/gkab1059 ).
Registering Samples
- Go to the EGA programmatic submission
- Submit a sample XML
- Save the list of accession numbers. These will be needed for the AF template.
Registering Data Access Committee
Further information on the role of your DAC.
- Go to the EGA programmatic submission
- Submit a DAC XML
- Take a note of the DAC accession number (EGACXXXXXXXXXXX)
Registering Policy
Your Data Access Policy provides the terms and conditions of data use. This is also referred to as the Data Access Agreement (DAA).
Completion of a DAA by the applicant/s should form part of the application process to the Data Access Committee (DAA)
- Go to the EGA programmatic submission
- Submit a Policy XML
- Take a note of the Policy accession number (EGAPXXXXXXXXXXX)
Complete the Array-based format (AF) spreadsheet
Once you have completed the registration of your Study, DAC and Policy using the programmatix submission, you must then complete and return the AF spreadsheet
The AF spreadsheet consists on four components:
Do not use EGA IDs registered using the Submitter Portal. You can easily identify objects registered in the Submitter Portal by their EGA ID pattern: EGA[A-Z]5{10 more digits} (e.g. for a sample EGAN50000002506).
For populating an AF spreadsheet, please exclusively use EGA IDs following this specific pattern: EGA[A-Z]0{10 more digits} (e.g. for a sample EGAN00001691542).
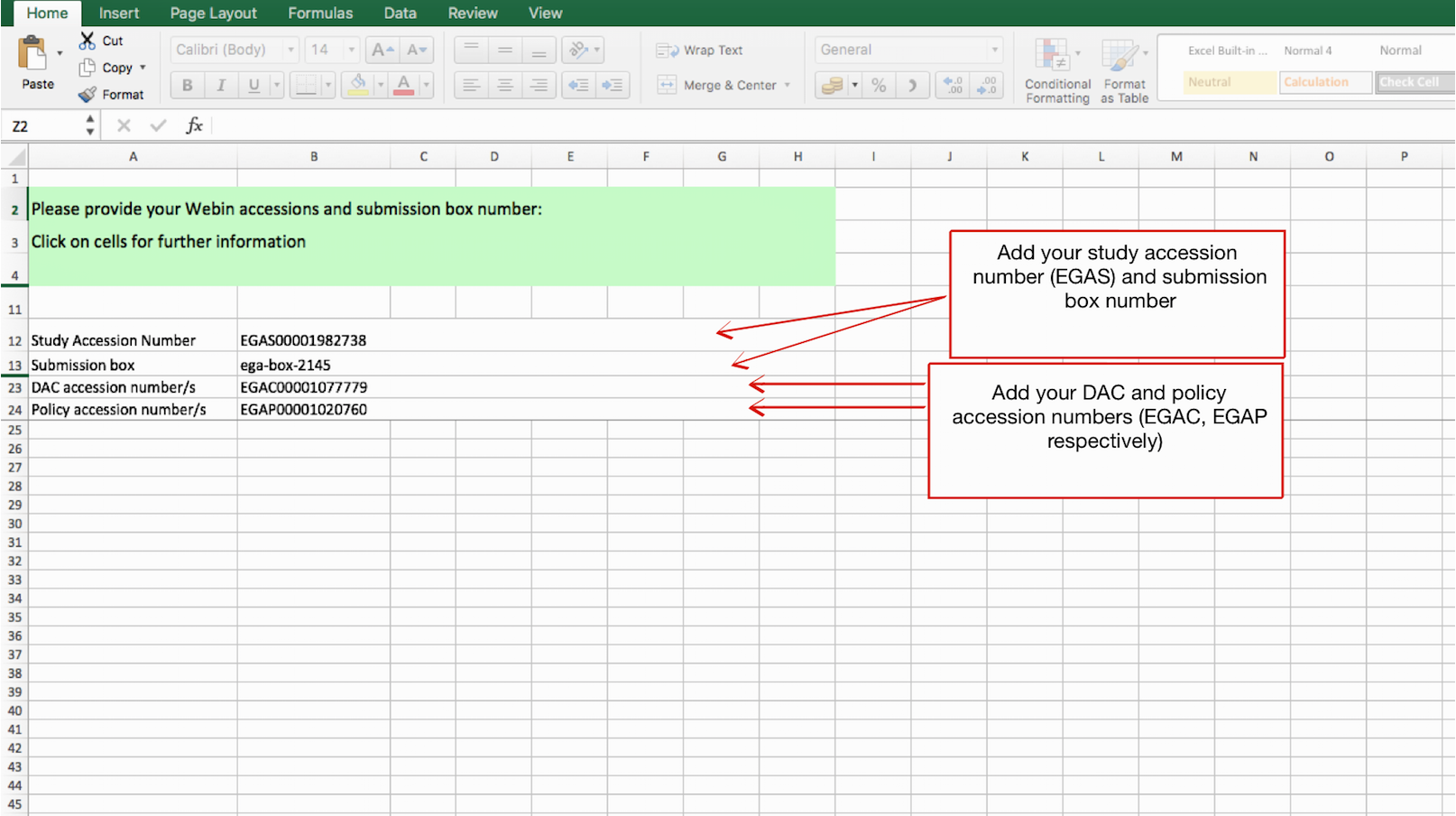
- Tab1) Webin accessions : Provide the accession numbers for your study, DAC and policy. Please, also add your ega-box number.
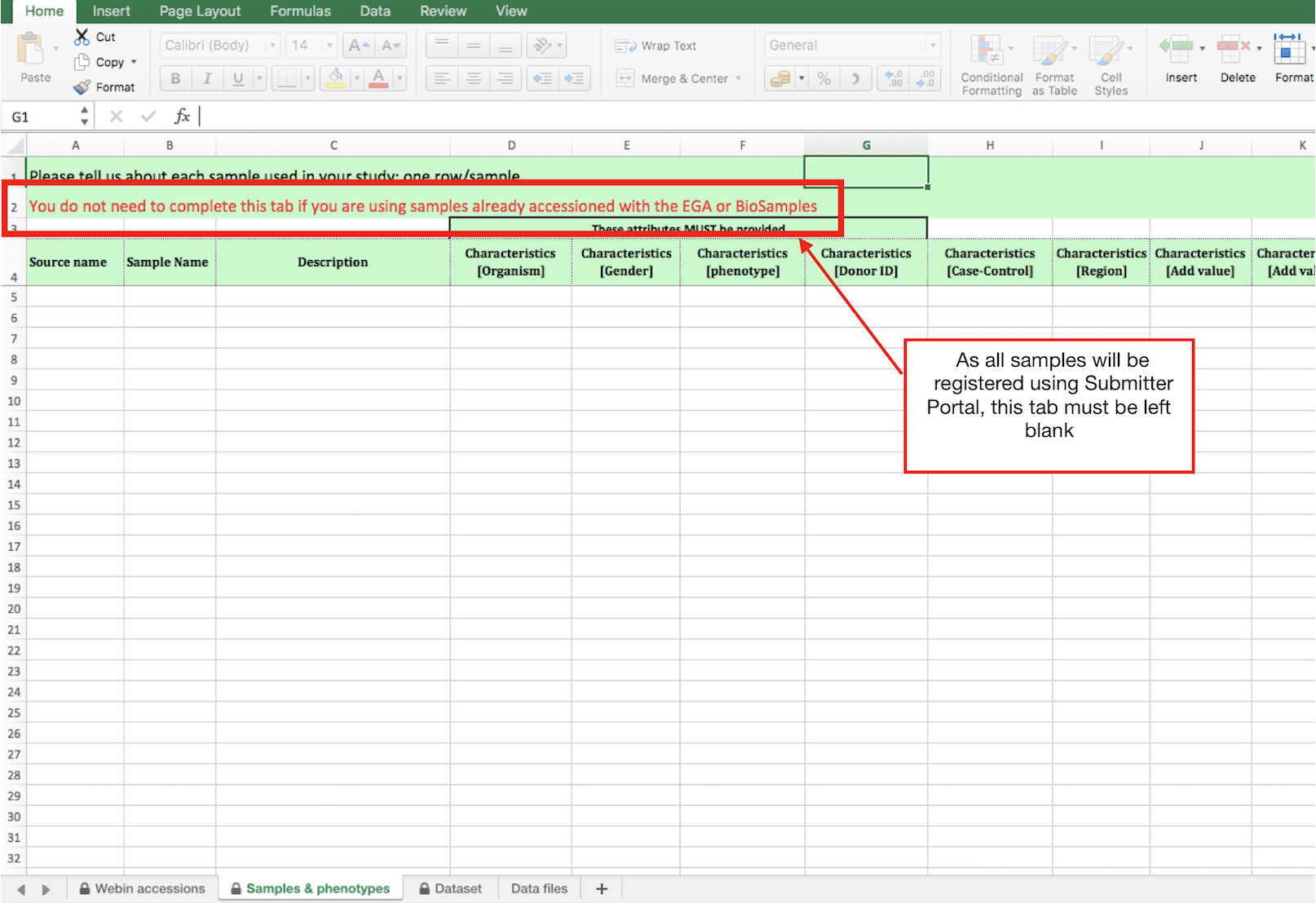
- Tab2) Sample & phenotypes: Please, leave this tab blank. All samples MUST be registered via Submitter Portal
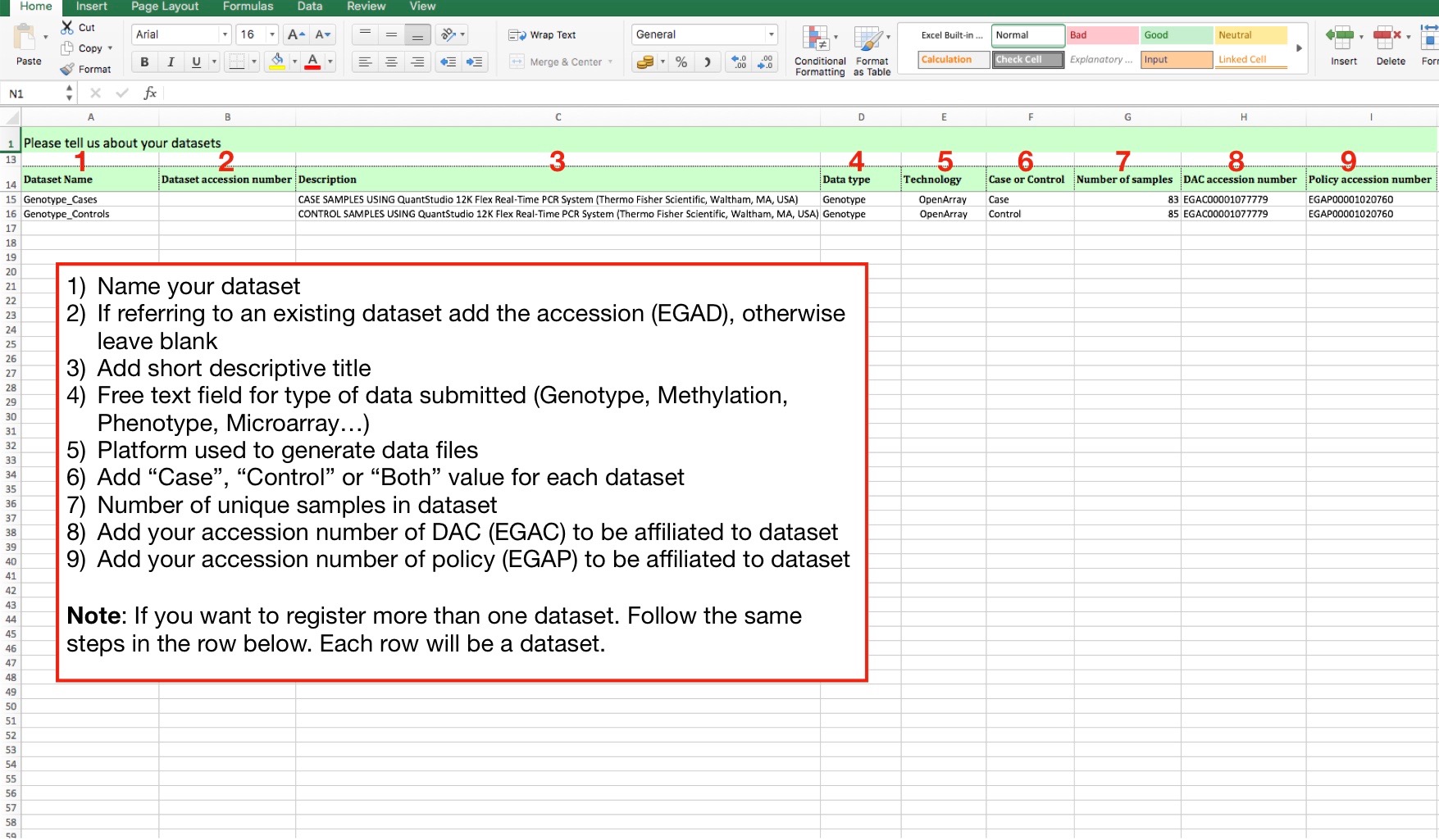
- Tab3) Dataset: Describe the dataset to be created
- Tab4) Data files: Define how your data is going to be organised into datasets and packets for distribution (linkage between samples and files).
Should further assistance be required after going through the guide below; please do not hesitate to contact the EGA helpdesk
Once the AF spreadsheet is populated, please send it to our EGA helpdesk for further validation.
AF spreadsheet
Should your submission require multiple DAC's or policies, use ' ; ' to separate the accession numbers.
AF spreadsheet: Samples & phenotypes
AF spreadsheet: Datasets
We suggest that each dataset consists of a common set of data. The example below consists of two datasets, grouped according to shared data type, technology and by case/control.
We also like to capture the number of unique samples that make up the dataset and the Data Access Committee (DAC) responsible for providing the named dataset and their policy (EGAP).
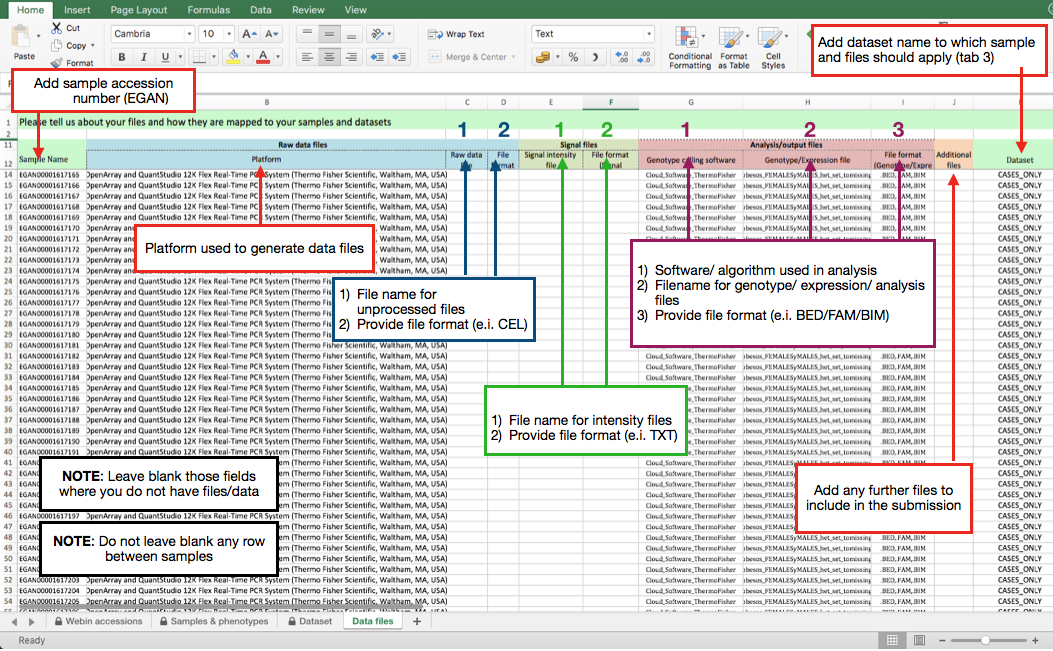
AF spreadsheet: Data files
What follows is an example of how to map your samples to the array based files added to your upload account (4th tab).
Please, find below some practical examples on how to register the linkage between samples-files
Case 1) 1 sample or list of samples in different datasets:
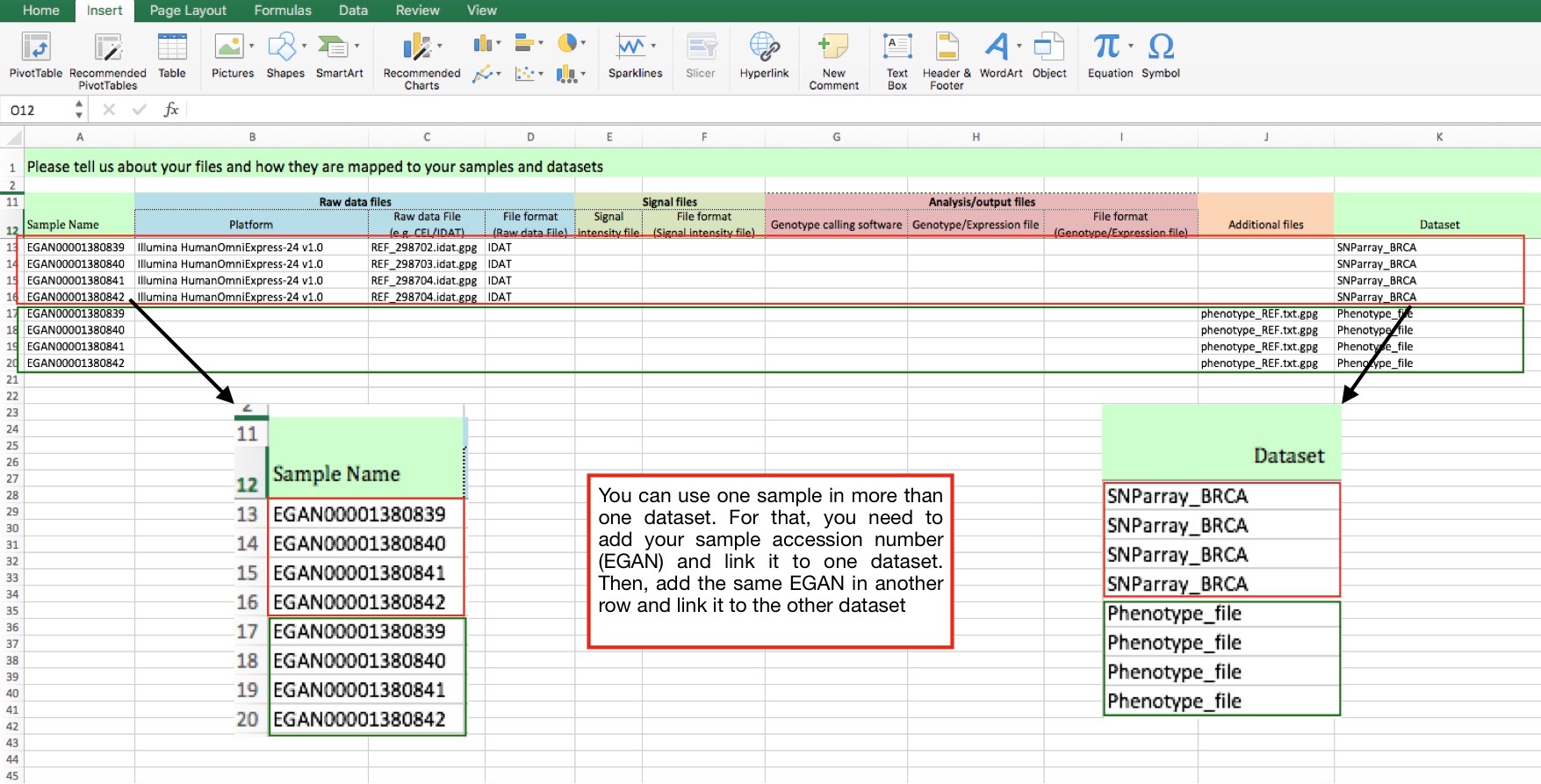
In case you have a list of samples that belong to different datasets, please, repeat the samples accession number/s in the first column and link the sample to the corresponding dataset each time (each row).
Each row is one linkage between sample-file-dataset.
Case 2) 1 sample links to several files:
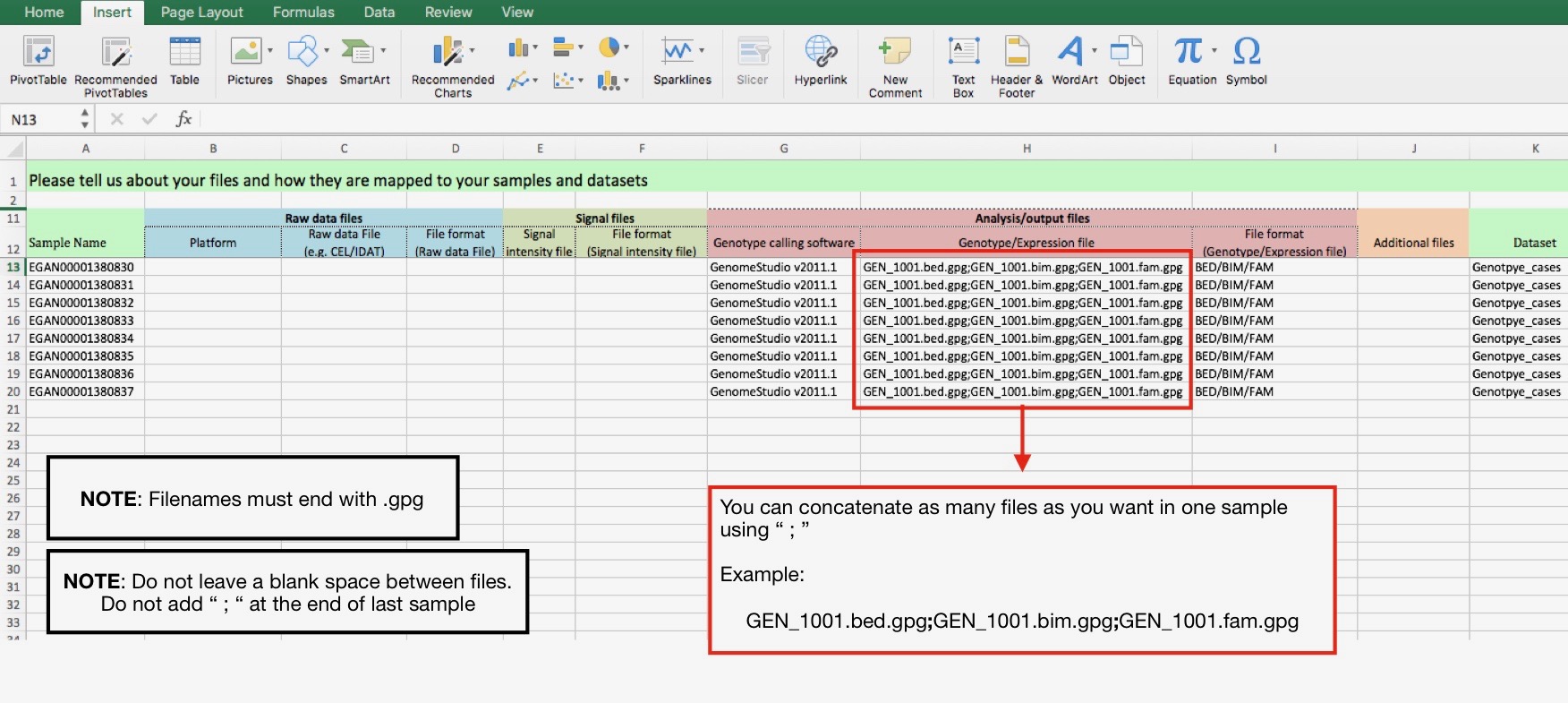
In order to add multiple files to one sample you MUST use “ ; “ between filenames. Example: file1.gpg;file2.gpg;file3.gpg
In case that you want to add an extra file to the sample (phenotype or .Rdata), please use “Additional files” column.
Important note: You MUST upload the encrypted and unencrypted md5sum values of all files uploaded to your submission account using the filename nomenclature (file.gpg, file.md5,file.md5.gpg). Your submission will not be processed without md5values supplied for all files in the CORRECT format.
What happens after the submission of a dataset?
All datasets affiliated to unreleased studies are automatically placed on hold until the authorised submitted or DAC contact instructs our EGA helpdesk for the study to be released.
Finally, your data is archived within our databases and prepared for encrypted distribution upon the request of permitted EGA account holders.
We strongly advise you NOT to delete your data until we confirm that your data has been successfully archived.
