Submission FAQ
Before Submission
Is the EGA the right archive for my data?
TThe most suitable archive for your data is dependent on the type of data you are wishing to submit and whether the data require public or controlled access. Public access is defined as complete and open access to all submitted data. On the contrary, controlled access, exerted by the EGA, requires formal applications to be made to access the submitted data files and metadata.
EGA only accepts human-derived data subject to controlled access. If your submission contains other types of data, please choose the appropriate repository for it (see image below): ENA, EVA, ArrayExpress, BioSD and GWAS catalog.
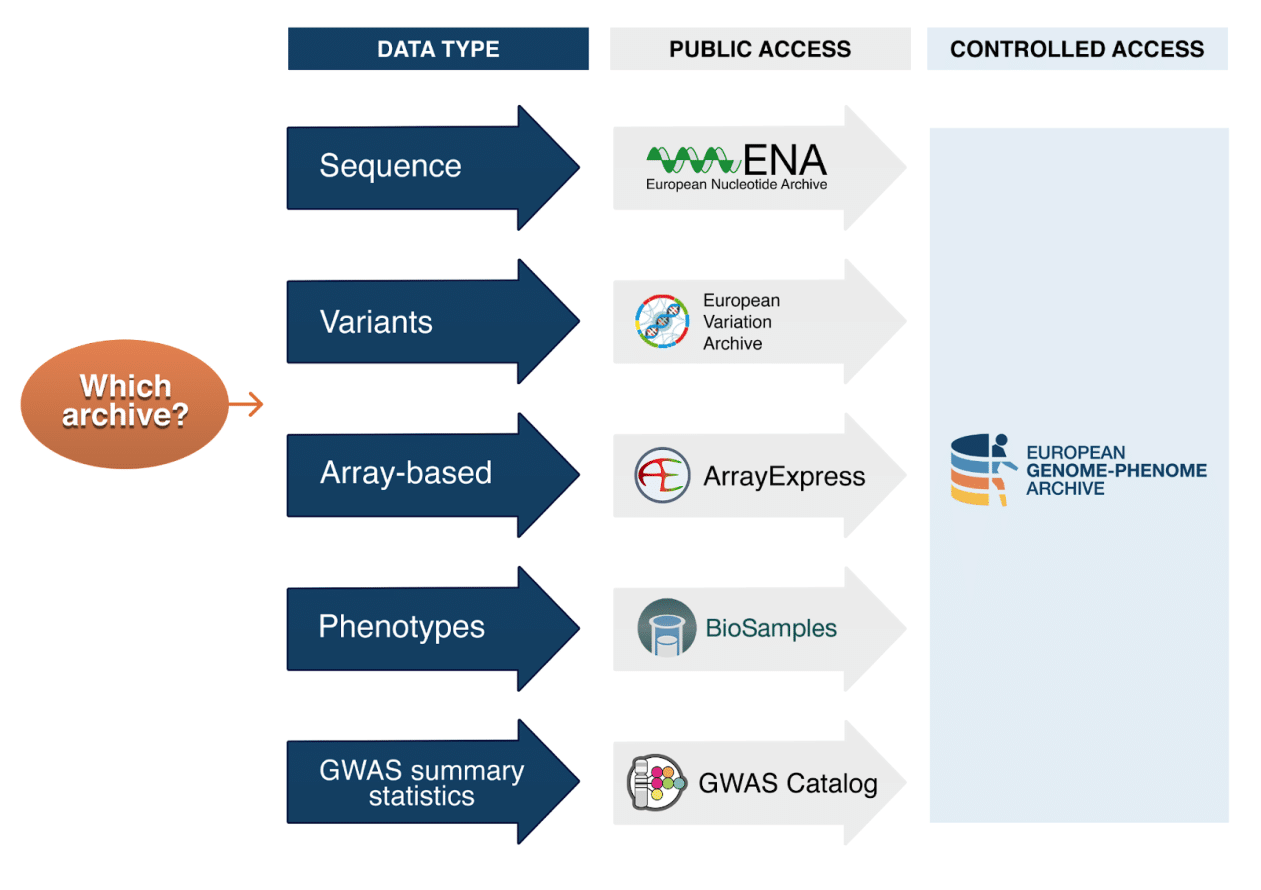
Should your submission be subjected to controlled access?
Data access conditions are normally defined in the original informed consent agreements signed by the participants involved in your study. All data submitted to the EGA is subject to controlled access. These consents prevent the derived data files, potentially identifiable, from being dispersed by open and public access. Controlled access data often refers to human data derived from medical research and consortium projects. If in doubt, consult the informed consent agreements that apply to your study
The EGA enables you to hold a submission before publication.
What data types can be submitted to the EGA?
Data types accepted by the EGA can be split into three categories:
- Sequences: both in generic and platform-specific formats.
- Array-based: from raw signal files to processed matrices.
- Phenotypes: all possible phenotype formats are accepted.
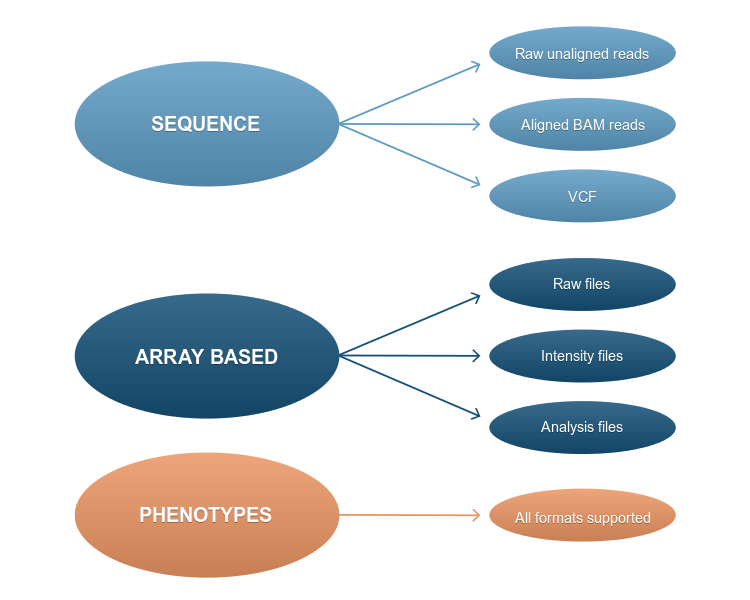
All manufacturer-specific raw data formats derived from major next generation sequencing platforms are accepted. Also generic sequence formats: flat reads in a FASTQ file, aligned sequences (BAM or CRAM files) as well as sequence variation files in VCF format.
All array-based technologies are accepted, including raw data, intensity and analysis files, without any restriction on data formats accepted.
We also accept and distribute phenotype data (associated to the samples) in almost any format: from an image to a README file.
How long does a submission take?
Submissions to EGA come in a variety of formats and sizes, thus it is difficult for us to exactly predict how long a submission will take. We, therefore, advise all of our submitters to allow as much time as possible to make a submission. Based on previous records, we anticipate that the submission process may take at least one month. The submitter’s familiarity with the procedures, possible technical issues that may arise during submission and the amount of data that is being submitted are the main factors that will affect the length of the submission process. However, each step of a regular submission should be considered when estimating the time it would take:
- Encryption of the files
- Upload of the files
- Metadata submission
- Archival of the files
- Release of the study and datasets to EGA webpage
For example, the upload of the files depends on the submission size, while metadata submission mainly relies on each submitter’s expertise. Further, some steps (e.g. answering to inquiries) depend on the EGA Helpdesk team, which may take some days to be processed during busy times
Is data deposited in the EGA secure?
The EGA set-up consists of a secure computing facility for data processing, a shared EBI set-up for data submissions and distribution of data via data requests made through the EGA website. Data is also copied in the Barcelona Supercomputing Center (BSC) infrastructure, where all stored and distributed data is encrypted
Data is encrypted along the submission process and stored securely, granting its access to authorised users exclusively. During the download process, through our Python Client or Aspera, all requested data is downloaded over secure https connections.
All data at the EGA is encrypted, and only accessible (for log-in and download) through secure protocols. For further information please, visit our security overview.
What documentation do I need to provide?
All submissions require policy’s documentation: 'Data Access Agreement (DAA)', 'Data Processing Agreement (DPA)' and 'Authorized Submitters Formulary'. The data processors (EGA) and the data owners will also sign the DPA.
Will all metadata be public?
Among the submitted metadata we need to make the distinction between identifiable and unidentifiable metadata: (1) the former may allow the identification of the human the sample derived from (e.g. detailed geographical providence, personal name, family ancestry…); (2) while the latter can be used to interpret the data without compromising the anonymity of the patients.
The majority of the metadata submitted to the EGA corresponds to the unidentifiable category (e.g. sequencer's model). This type of metadata is publicly available on the EGA website and other EBI resources/partners’ websites.
On the other hand, some parts samples’ metadata are subject to being identifiable, and thus only accessible by authorized data requesters, with the exception of:
- 5 submitter-defined attributes of the sample: alias, title, subject_id, gender and phenotype. It is the submitter’s responsibility not to submit sensitive metadata in these public fields.
- 3 anonymised fields that pinpoint the sample record in archivals: sample’s EGA stable ID (EGAN…), BioSample ID (SAMEA…) and submitter’s center name.
During Submission
Are there any sample specific requirements for EGA?
All samples submitted to the EGA must include the attributes of biological sex, subject ID (anonymised individual identifier) and phenotype information. These are critical for data findability and its analysis, and we highly recommend using controlled ontology terms where applicable. For example: defining tumour and non-tumour samples and/or defining disease state.
The EGA recommends using the Experimental Factor Ontology Database to find ontologized terms that describe your sample phenotypes.
How do I get an accession number to use in my publication?
You will receive your study accession number (EGAS…) upon complete your submission, either:
- Programmatically. As soon as the metadata is submitted and validated your study will be assigned an accession number that will be given in the submission’s response.
- Manually registering your study and relevant metadata using the online metadata submission tool: the EGA submitter Portal.
How are files uploaded to the EGA?
Data files are uploaded into private submission drop boxes (i.e. environments to which you are granted access and where you can transfer your files) using INBOX or FTP. These spaces are provided as part of the submission procedure. Before uploading any file, you must encrypt your files, . Only encrypted files shall be uploaded to the drop boxes.
Why does data need to to encrypted for my submitted files?
It is one of the security steps the EGA has implemented. In case of a security breach, people without the proper encryption key will not be able to read or use the information that could have been leaked. This measure is essential when working with sensitive data, such as controlled access human data.
All submitters must use crypt4gh to create EGA compliant files prior to uploading them. This encryption is GPG-based, using EGA’s public key.
Why are my files not available if I see them in the INBOX?
There exists a time window between the data upload and the availability of such files via the Submitter Portal. For this reason, some metadata (run and analysis objects) cannot be registered until at least 24 hours after the files have been uploaded to your box.
Why are MD5 sum values generated for my submitted files?
We require pre- and post- encryption MD5 (message-digest) checksum values to be provided for all submitted files. These 128-bit values are computed using the content of each file, creating unique sequences that allow us to ensure that file integrity has been maintained during the transfer process. In other words, if the MD5 checksums we generate and those you generated match, we infer that the content of the transferred files is correct (i.e. files are not corrupted or truncated). MD5 checksums are computed automatically using the crypt4gh tool provided.
Your submission will not be accepted and may be significantly delayed if you do not provide MD5 checksum values for all data files in the required format.
How can I check if my files are correctly uploaded to the inbox?
It is important to check the status pf your file so you know whether your files are in the inbox, being processed, or if there is any issue with one of them. In order to check this, you should:
- Look for the file locally.
- Drag and drop it to the file table. Then bars will appear on the table, which means that we are processing it.
- Green: The files checksum are correct and your file will move to “ingested files”. No further actions are needed from your end..
- Red: The files checksum does not match and your file needs to be re-uploaded. Please re-upload relevant files to your inbox using the same path.
After Submission
How do I use my accession number in my publication?
We suggest the use of the below template, using your study accession ID (EGAS…) :
Data has been deposited at the European Genome-phenome Archive (EGA), which is hosted by the EBI and the CRG, under accession number EGASXXXXXXXXXXX. Further information about EGA can be found at https://ega-archive.org and "The European Genome-phenome Archive of human data consented for biomedical research"
Your study ID will be the one that groups your whole submission, and thus its usage is recommended as such. Nevertheless, all metadata submitted to EGA hold a unique and persistent identifier (starting with EGA…) that can be used to identify specific records. For example, you could reference a specific dataset (EGAD…) or sample (EGAN…) in your publications (see full list of identifiers).
How do I make my data searchable?
Once you have finalized your submission, you can schedule the data release. Please take into account that the release process needs time for the files to be archived in our system, and for the Helpdesk team to validate your submission.
Can I withdraw (meta)data from the EGA?
We have methods in place for the secure removal of deposited (meta)data. Contact EGA-helpdesk for further details.
EGA complies with FAIRness of (meta)data, and thus, even when the data is removed we keep an entry for their identifiers in our system. In other words, we execute a soft delete on canceled objects (e.g. a study): metadata is still stored in our systems, but it loses all links, cannot be queried and data files cannot be retrieved anymore. The reason behind this behaviour is so that queries using withdrawn data properly respond back (see example of a canceled study).
What happens to the data once it has been submitted to the EGA?
When the data is submitted, the submitter can choose either keep their data private or schedule the release of their data.
