Submitter Portal
The Submitter Portal does not support array submissions! Please refer to the array documentation
Any new objects registered in the Submitter Portal will not be available for use in an array submission
For further information please check our Submission FAQs, submission quickguide as well as submission terms!
The metadata submission process can be difficult and time-consuming. For this reason, the EGA has developed the Submitter Portal, a tool that was created to offer a simplified and user-friendly method of registering metadata.
Our portal provides features that are intended to make the input of metadata easier, ensuring that your data is registered correctly and effectively. Our page is divided into logical sections and includes a helpful video instruction to further assist you as you complete the submission process. With the Submitter Portal, you can rest assured that the submission of your data is in good hands.
Submitter Portal Index
Previous steps
Create your EGA account
To submit data, you first need to create your EGA user. Then, once your account has been verified by the Help Desk team, you will be able to request a submitter role and send the EGA Data Processing Agreement (DPA).
In the case you already have an EGA account, or an ega-box (submission account), you can skip this step.
Upload your files
Please note that all your files must be encrypted using the Crypt4GH tool before upload them.
As soon as you are assigned with a submitter role and added your public key to your profile, you will be able to connect to the EGA inbox and upload your files.
Understand the EGA metadata schema
It's crucial to comprehend the EGA metadata schema, a set of rules that specify how data is organised, described, and shared inside the EGA, in order to get the most out of this resource. Learn all about EGA metadata schema!
Register your DAC and policy
There are two objects in the EGA metadata schema that are registered in a separate portal. All DACs and policy objects are registered using the DAC Portal, a tool developed by EGA to help data controllers manage their data stored at the EGA. You can find relevant information in the DAC Portal Guide.
Access to Submitter Portal
1. Request Submitter Role
Once you have created your EGA user, request a Submitter Role and complete relevant information such as submission type and size.
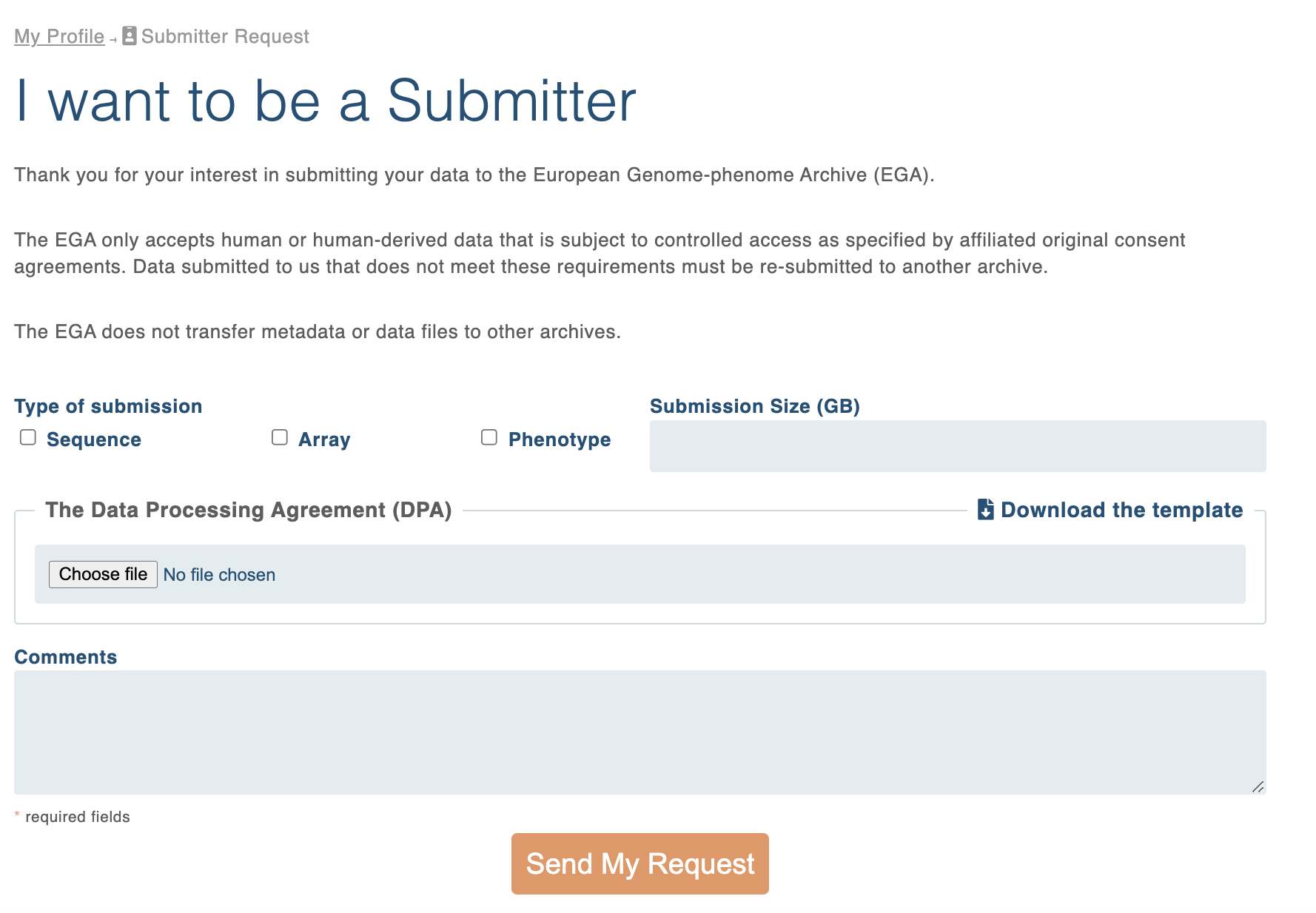
2. Signature of the Data Processing Agreement
The Data Processing Agreement (DPA) signed by CRG and EMBL-EBI (Joint Data Processors for the EGA) can be downloaded to the submitter for its signature. Further information related to the DPA can be in the Privacy Notice section.
The DPA should be signed by a legal representative of the submitter with power of attorney/appointment duly authorised to sign contracts on behalf of the institution. The submitter will return a fully-executed copy of the DPA to us. Please note that due to the high volume of submissions to EGA the DPA is non-negotiable. Once you have completed all the information and attached the DPA signed, send your Submitter request and the EGA Helpdesk Team will validate it and send a confirmation to start submitting your metadata.
Start a Submission
The steps below will show you how to start a submission and are accompanied with a tutorial and the Take the tour of Submitter Portal.
1. Login
Go to the Submitter Portal interface and log in using your EGA account credentials.
2. Create a submission
The Submitter Portal interface will show you any already existing open submission. To start a new submission click on “Create a Submission”.
In this phase you have the option to add any collaborator(s) as well as their permissions (“Read only” or “Read & Write”). If you want to invite all the collaborators from a previous submission, you can add them by selecting said submission.
In the event that the submission has been closed, you will need to reopen it. To do this, you will need to modify one of the objects in the submission, for example, copy and paste the title. Once the submission is open, you can add a collaborator.
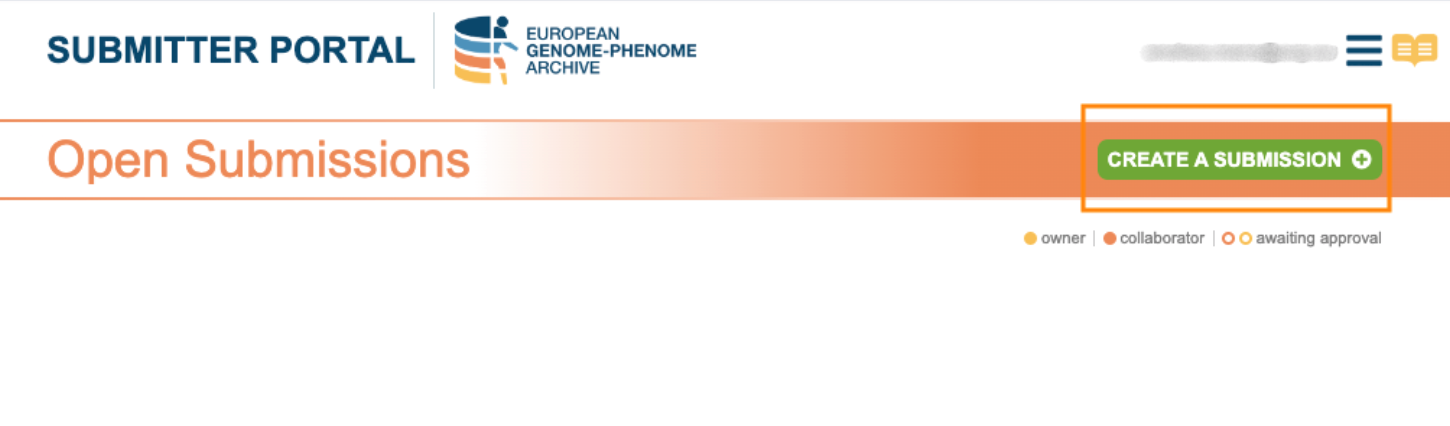
Further information can be found in the Submission Tour
3. Create Study
Click on add Study and complete the form with the relevant information. The fields with an asterisk (*) are mandatory.
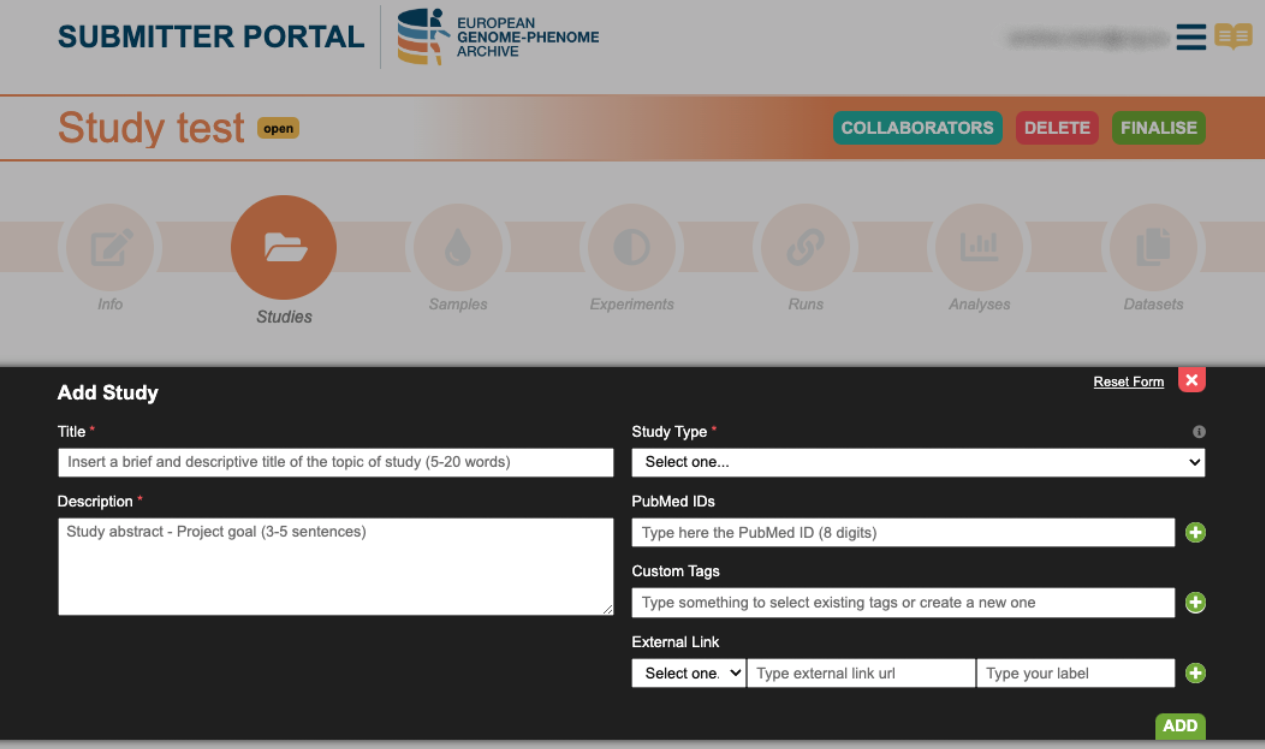
Further information can be found in the Submission Tour
4. Create Sample
Sample registration can be done either individually or in batch by using a template.
- Batch upload: download the template and complete it with information about the sequencing samples. Upload the samples.csv and the sample will be created.
- Add single sample: complete the mandatory fields with the metadata information of your sample.

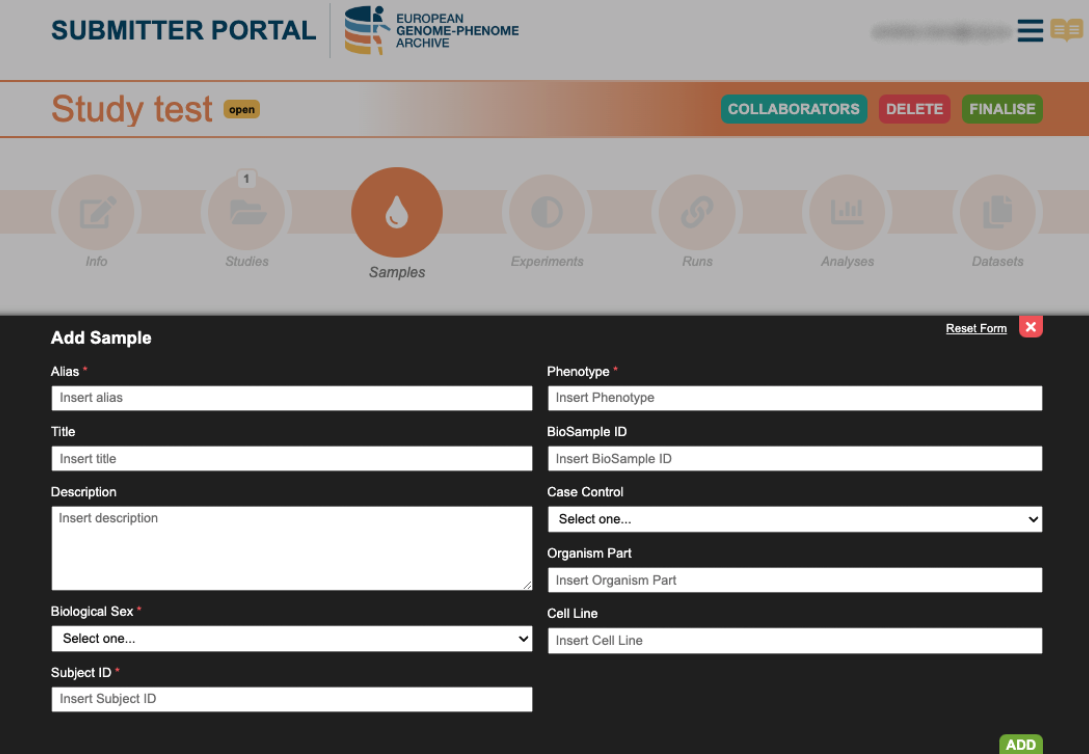
Further information can be found in the Submission Tour
5. Create Experiment
Register an experiment with information about the sequencing methods, protocols and machines. Experiments generate the connection between samples and study.
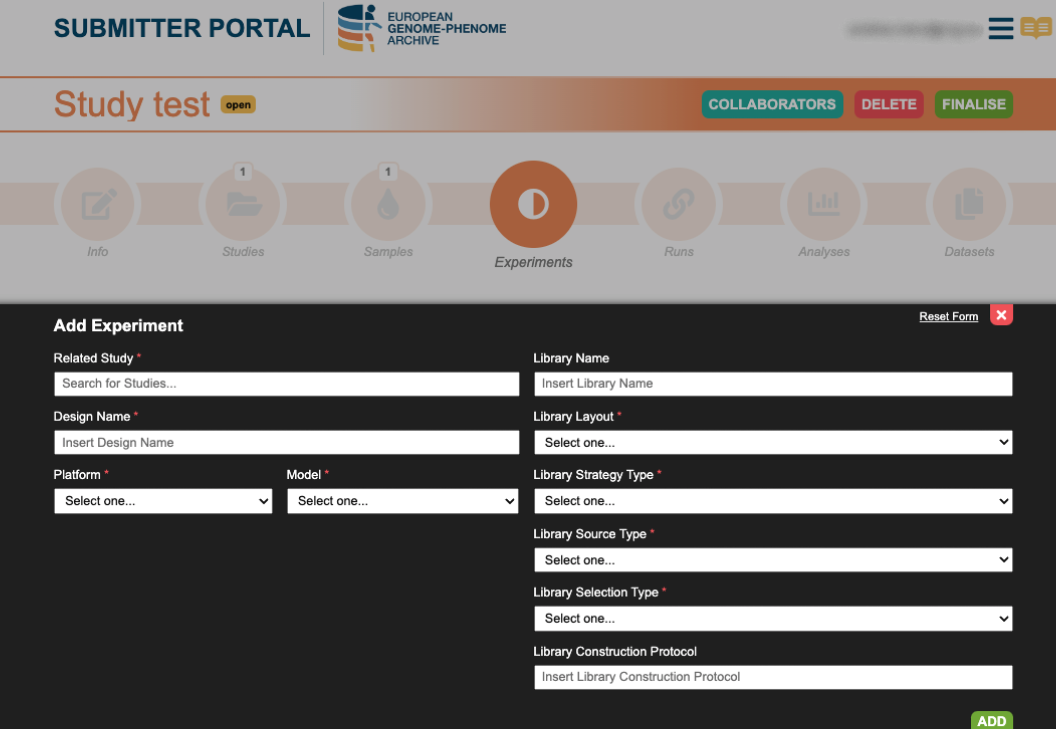
Further information can be found in the Submission Tour
6. Create Run
Samples, experiments and files are linked through runs. To register a run, you will be asked to select the appropriate experiment and the file format. Run registration can be done either individually or in batch by using a template.
- Batch upload: download the template and complete it with the sample - file linkage. Upload the run.csv and the runs will register.
As the INBOX is a directory, bear in mind to include the file path (/). Example: /filename.c4gh. - Add single run: select the file format, sample and experiment from the drop box.
For re-using submitted samples, please use the EGA accession ID (EGANXXX).
Please note that files must be uploaded in your inbox before linking your files and samples.
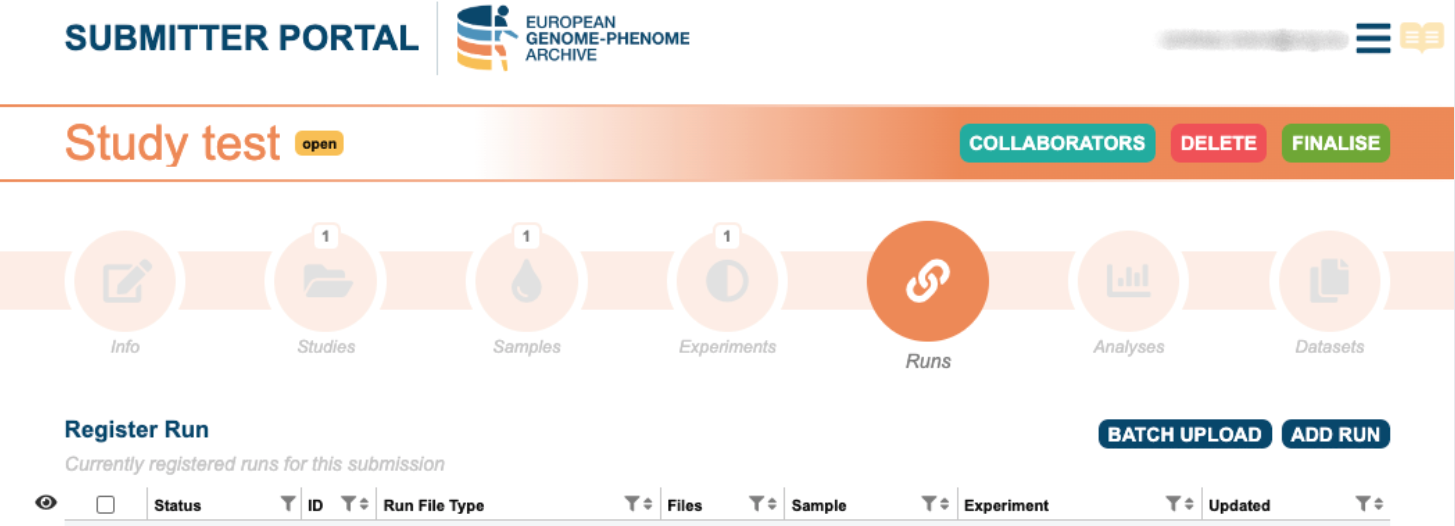

Further information can be found in the Submission Tour
7. Create Analysis
Analysis should be only used for BAM/BAI or CRAM/CRAI pair, VCF and phenotype linkage to samples.
The analysis is an EGA specific metadata object that links samples to files. This object also stores some metadata about your experiments, such as the experiment type, genome reference, or the platform used.
**If only BAM or CRAM alignment files are submitted but not the original unaligned FASTQ files, then please make sure that the BAM or CRAM files also contain the unaligned reads. This is critical to enable primary re-analysis and re-alignment of the dataset using new tools or future genome assemblies.**
Start by selecting the samples to be linked to the file, and then populate the required attribute fields. Please be aware of the existence of mandatory fields that must be populated.
When populating the chromosome field (mandatory), please select the chromosomes from the checkbox.
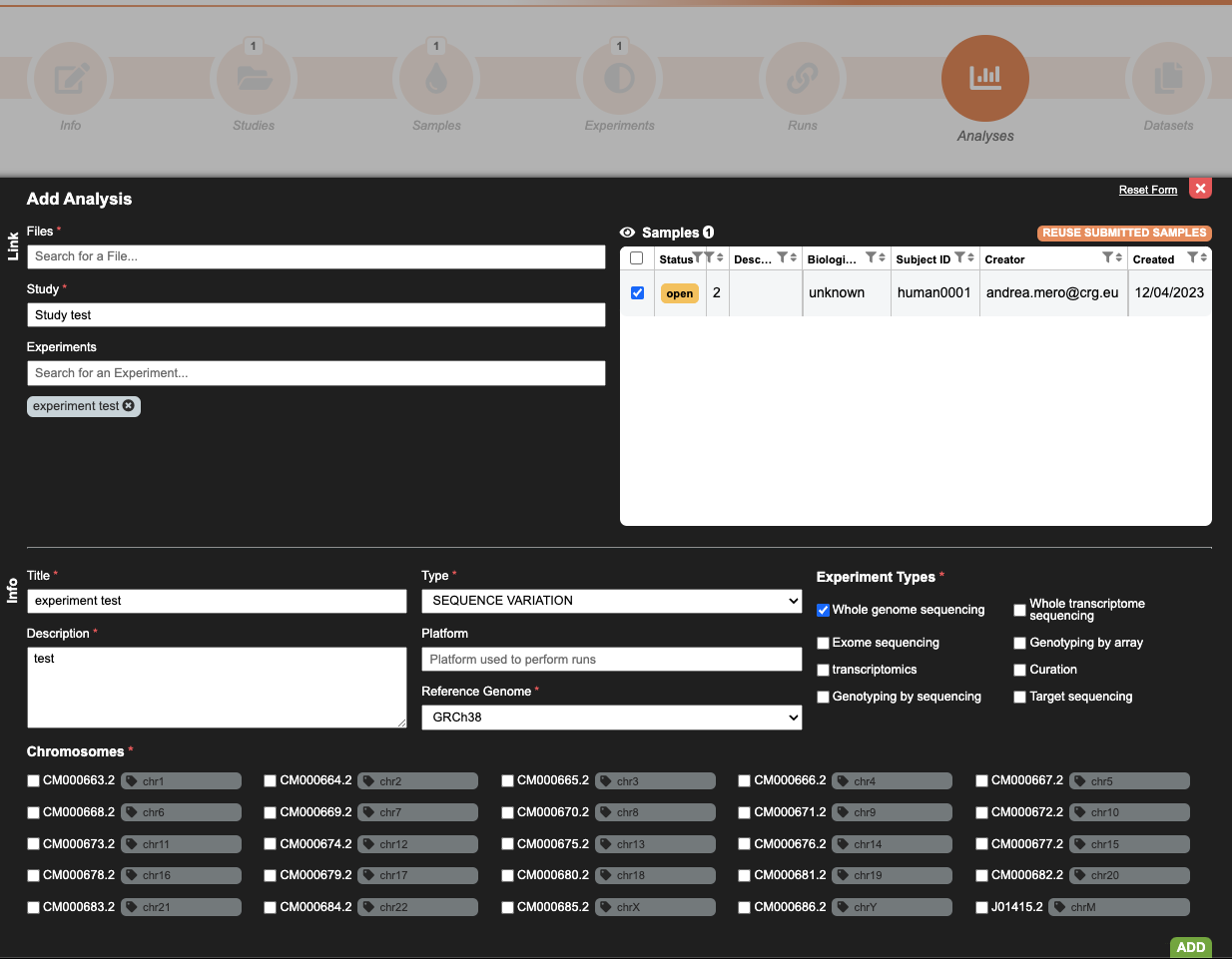
Please take in consideration that EGA allows for the re-use of any registered metadata. Therefore, a previously registered study or samples can be referenced for the analysis in the current data submission.
Further information can be found in the Submission Tour
8. Create Dataset
Dataset contains the collection of runs/analysis data files subjected to controlled access by being associated with a Policy.
Complete the mandatory fields such as title, description, type and policy. Please, take in consideration that DAC and Policy must be registered in advance in the DAC Portal.
Similar to other metadata objects, you can re-use previous runs or analyses, using their EGA accessions IDs (runs - EGAR, analyses - EGAZ).
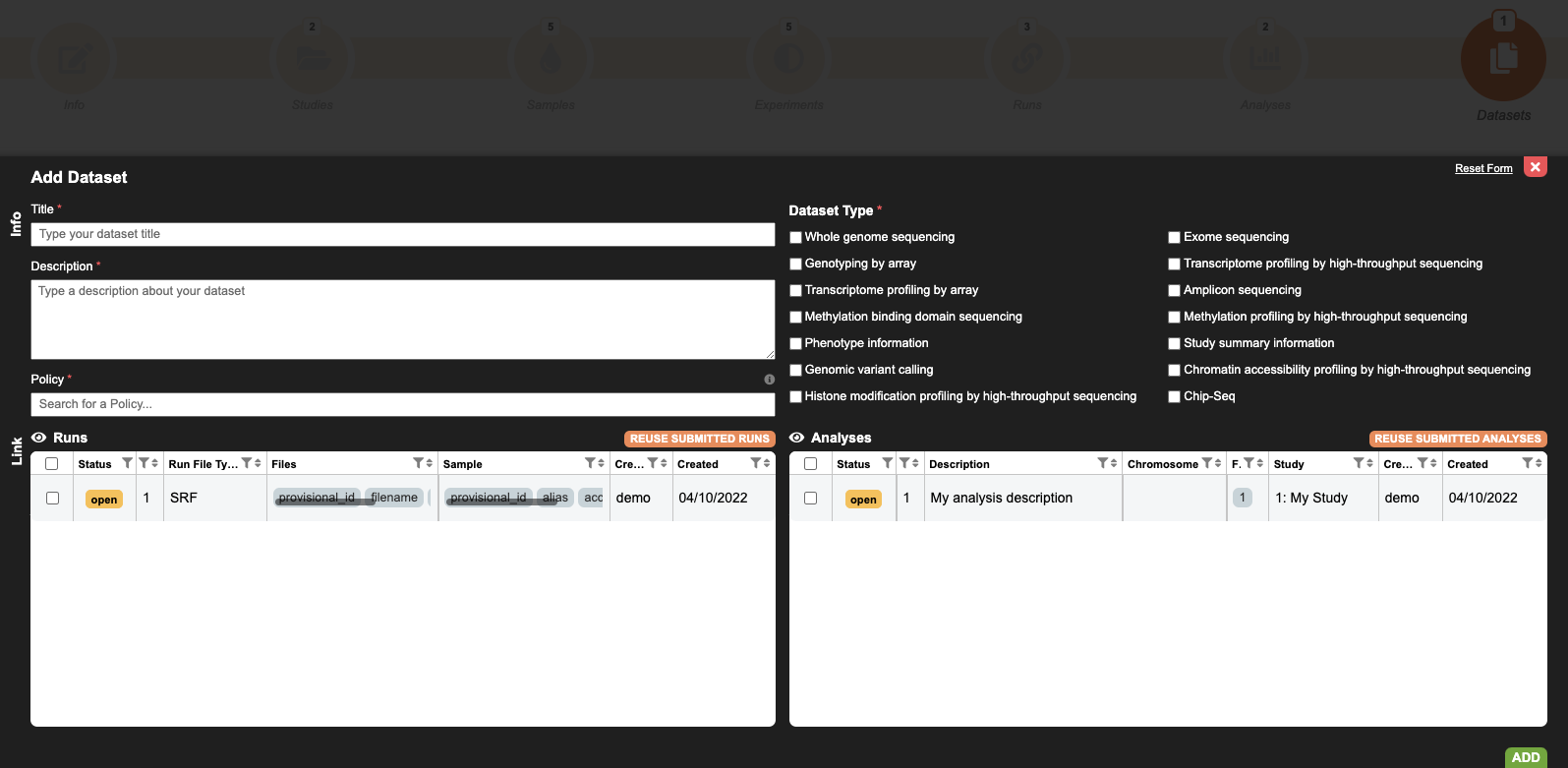
Further information can be found in the Submission Tour
9. Finalise submission
Once the files are ingested please delete them from your SFTP INBOX before finalising your submission. This will prevent the files from reappearing in your Submitter Portal Inbox.
Once you have completed your submission, click on the Finalise button, and schedule an estimated release date. Afterwards, your submission will be sent to Helpdesk for approval.
Please note that once the Helpdesk has approved your submission, you will receive the identifiers for your data. See the Frequently Asked Questions (FAQ) for more information.
10. Update closed submission
If you want to append more file to a closed submission, you will need to re-open the submission where the dataset was registered.
- Find the Submission: go to the Submitter Portal Menu -> Datasets section and find the submission id (EGA0XXX).
- Reopen the submission where the dataset is located: to reopen a submission, you must edit any object within the submission. For example, you can copy and paste the title.
Take The Tour
The Submitter Portal Take The Tour is a visual guidance of all the steps described previously about how to start a submission.
Tutorial
In this tutorial we will show you how to use the Submitter Portal to register your metadata. Please note the video focuses on the run-based submission (for raw files - fastq - and aligned data - BAM/CRAM) and analysis-based submission (for your BAM/BAI pairs, variation -VCF - and phenotype files).
